You aim:
- To know the genetic predispositions of your pet to certain diseases
- To confirm the colour of your foal's coat or to check the presence or absence of horns on your calves as part of a breed improvement
- To check the presence of an insert in a plasmid
- To look for the presence of mutations involved in antibiotic or antifungal resistance mechanisms in microorganisms
- To identify the sequence of one or more target genes for filiation or genotyping studies
To perform a backup? of the sequence of an antibody of pharmaceutical interest....
Sanger sequencingSequencing by the Sanger method, which allows to determine the base sequences of short DNA fragments, is one of the techniques commonly used to meet these needs.

This DNA sequencing technique, also known as "chain termination sequencing" or "deoxy sequencing", was developed by the biochemist Frederick SANGER in 1977 and involved two steps:
- A specific PCR: it differs from a conventional PCR using a single primer (instead of two) and by the incorporation of chemically modified nucleotides blocking the elongation of the fragment, the dideoxynucleotides (ddNTPs). The presence of a single primer prevents the amplification of the fragment to be sequenced. The ddNTPs, on the other hand, allow to obtain fragments of different lengths. The concentrations of ddNTPs used make it possible to statistically synthesize all fragment lengths. In the basic method, 4 reactions were performed simultaneously, each including a specific ddNTP (ddATP, ddTTP, ddCTP, ddGTP). Nowadays, each of the 4 ddNTPs carries a specific fluorochrome, which allows all DNA fragments to be labelled at their ends during a single reaction.
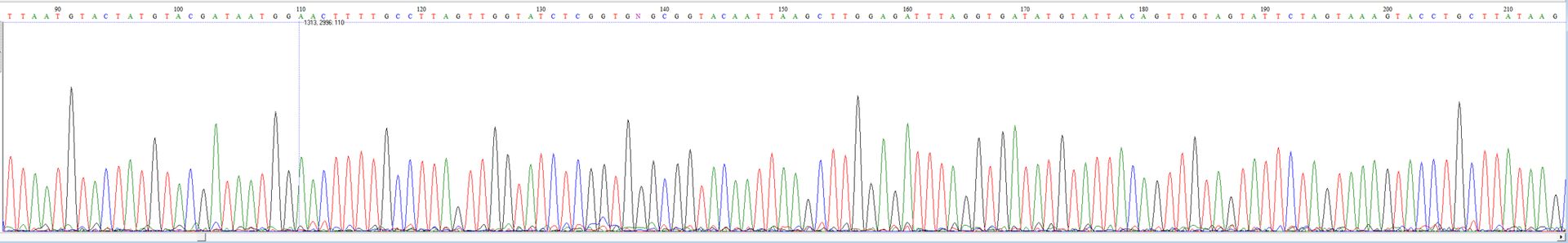
- Fragment separation: in the original method, the products of each reaction were separated by electrophoresis trough distinct agarose gel tracks. As the fragments were separated according to their length, it was sufficient to "read" the bands on the gel to determine the sequence. Nowadays, the DNA fragments obtained are subjected to capillary electrophoresis during which the fluorescent signal emitted by each labelled ddNTP is analyzed by computer. Data processing allows to determine the identity of the nucleotide and to reconstruct/reconstitute the sequence of the original DNA template.
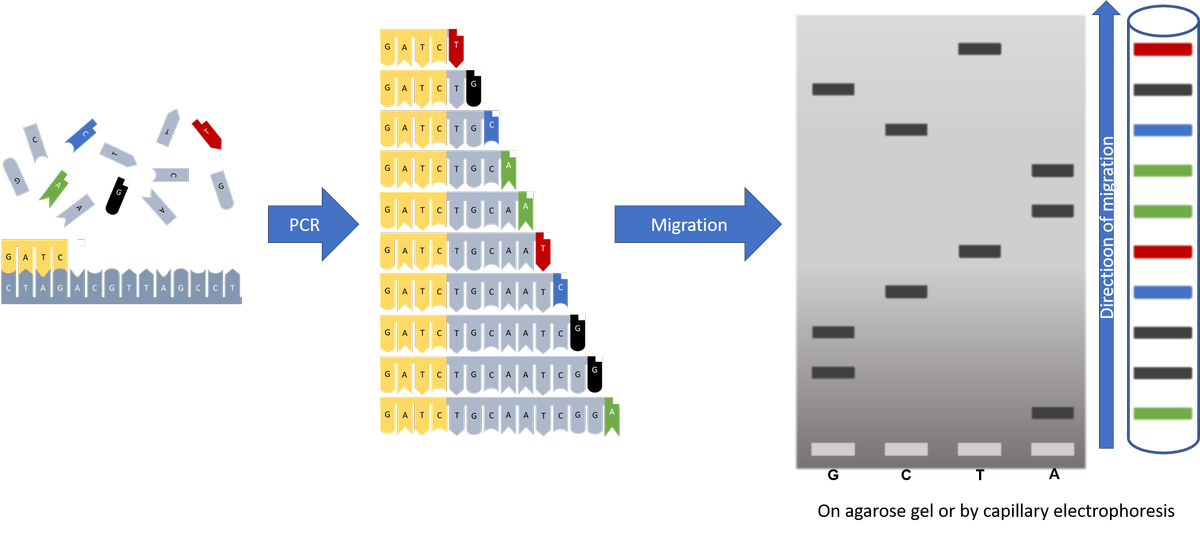
Migration profile after capillary electrophoresis

Biomigene is equipped for the realization of your sequencing rections with a SeqStudio device (Applied Biosystems).
Your samples can be supplied in different formats (PCR plates or tubes). For an optimal quality of the sequences, a quality control of your samples is carried out.
Sequencing is performed within 72 hours and the encrypted data is provided in.ab1 format and/or in FASTA format (text file) by e-mail.
For complex matrices such as DNA fragments rich in G/C bases, we propose an adapted protocol to reduce the formation of secondary structures that inhibit the PCR reaction.
Samples and sequences are kept in our premises for a period of 2 months, unless specified indications.



