
You wish :
- to determine a sequence to identify mutations (point mutations (SNPs), insertions, deletions)
- to evaluate the relationship between individuals or populations
- to highlight the presence of a specific organism in a biological sample
- to insert a gene into an expression vector for the synthesis of a recombinant protein ...
These analyzes generally require an amplification of genetic material by an enzymatic reaction called PCR (Polymerase Chain Reaction). This reaction is usually carried out from DNA. For certain specific applications, cDNA obtained by reverse transcription of RNA is used as starting material.
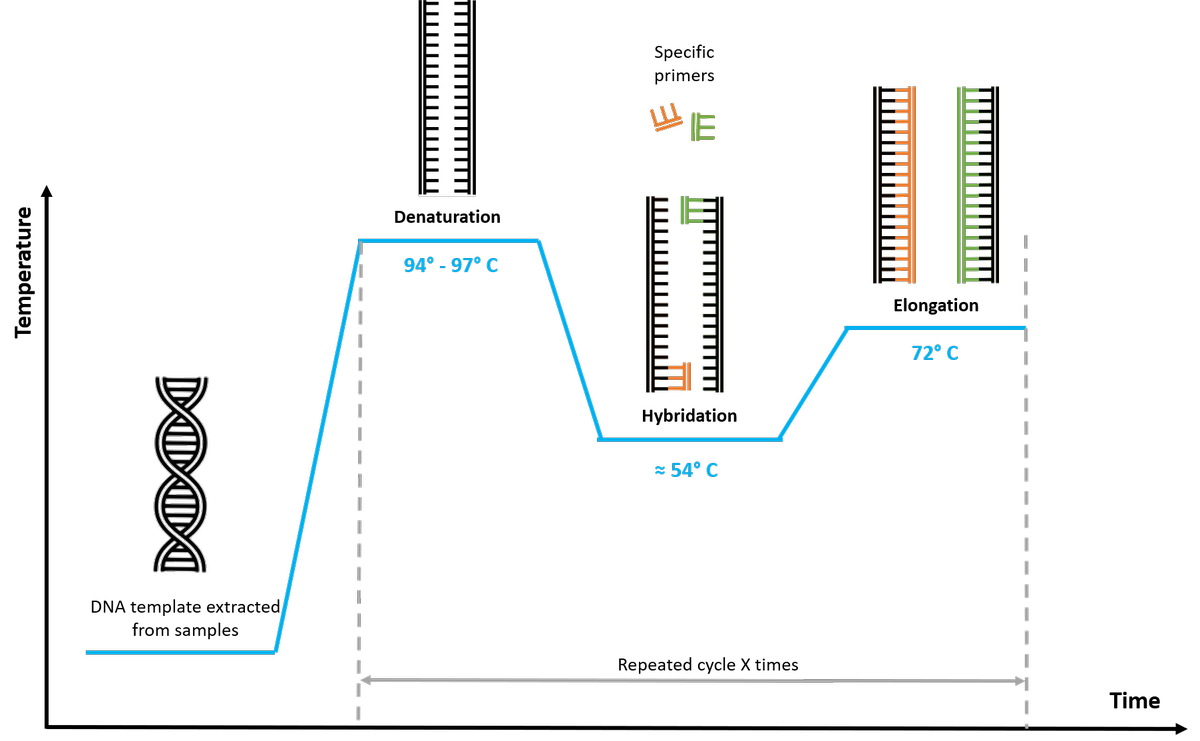
DNA molecules, in their natural state, are composed of two complementary strands. An increase of temperature above 94 ° C results in the separation of the two strands of the DNA and the destruction of the secondary structures (denaturation step).
The single-stranded DNA molecules are then paired at their ends to small fragments called primers during the hybridization step taking place at a temperature of about 54 ° C (depending on the base composition of the primer).
During the elongation step, an enzyme called DNA polymerase that optimally functions at 72 ° C, elongates these primers to form the strands complementary to the original template thus generating two double-stranded molecules identical to the initial molecule.
A new cycle (denaturation - hybridization - elongation) can then begin.
The PCR reaction consists of N cycles (generally between 30 and 40) leading to the synthesis of 2 N copies of the original template.

For best results, PCRs are performed using reagents (Enzymes, buffers, ...) adapted to your needs.
For example, for issues requiring to know precisely/the nucleotide sequences such as the search for SNPs (single nucleotide polymorphism) or spontaneous mutations, we will use a high fidelity polymerase.
For issues based on the size of the amplified fragments, the use of a conventional polymerase is sufficient.
Specific protocols were also developped for the difficult amplification of G/T and G/C rich fragments. Program (temperatures, step duration and cycle number) and composition of the reaction mixture are adjusted according to the size and base composition of DNA template and primers.
At BIOMNIGENE, PCRs are performed on Mastercycler X50s thermal cyclers (Eppendorf). These devices are equipped with a system that allowing to test an unequaled number of temperatures and thus, quickly determine the optimal conditions for the amplification of your fragments.
Upon requests, we carry out the purification of your PCR products. Concentration and quality of your amplicons can also be evaluated by measuring the absorption in the UV with a nano volume spectrophotometer or by a fluorometric assay on Qubit.
A report containing the amplification conditions, concentrations, relative purity of your amplicons will be provided.
The amplified genetic material may be returned to you by carrier or used by Biomnigene for further applications (fragment analysis, cloning, sequencing).




